In this post I have a few goals:
1. Become (re-)familiar with available geoms
2. Become (re-)familiar with aesthetic mappings in geoms (stroke who knew?)
3. Answer these questions:
- How often do various geoms appear and how often do they have required aesthetics?
- How often do various aesthetics appear and how often are they required?
- What geoms are most similar based on mappings?
The Back Story
Two weeks go I made whipped cream for the first time. In a tweet I lamented not having tried making it earlier in my life:

This was an error that resulted in missing out because of a lack of confidence. Now the opposite tale. Missing out because of false confidence. I know ggplot2…there’s nothing new to learn. Recently I realized it’s been too long since I’ve re-read the documentation. I’m betting my time making this blog post that there’s a few more like me.
I’m teaching an upcoming analysis course. To prepare I’m reading and rereading many important texts including R for Data Science. In my close read I noticed that some ggplot2 functions have a stroke aesthetic.

Didn’t know that…I figure I needed to spend a bit more time with the documentation and really get to know geoms and aesthetics that I may have overlooked.
What’s an aesthetic Anyways?
ggplot2’s author, Hadley, states:
In brief, the grammar tells us that a statistical graphic is a mapping from data to aesthetic attributes (colour, shape, size) of geometric objects (points, lines, bars)….aesthetics…are the properties that can be perceived on the graphic. Each aesthetic can be mapped to a variable, or set to a constant value. pp. 4; 176
So without aesthetics, geoms can’t represent data.
Getting the Geoms and Their Aesthetics
Before getting started note that wordpress.com destroys code. To get the undestroyed version of code from this blog use this gist: https://gist.github.com/trinker/a03c24a7fe9603d3091f183ffe4781ab
I wasn’t sure how I was going to accomplish this task but a little digging showed that it was pretty easy. First I used Dason Kurkiewicz & I’s pacman package to get all the geoms from ggplot2. This is where I first hit a road block. How do I get the aesthetics for a geom? I recalled that the documentation tells you the aesthetics for each geom. I noticed that in the roxygen2 markup the following line that creates the aesthetics reference in the documentation:
@eval rd_aesthetics("geom", "bar")
That allowed me to follow the bread crumbs to make the following code to grab the aesthetics per geom and a flag for when an aesthetic is required:
if (!require("pacman")) install.packages("pacman")
pacman::p_load(pacman, ggplot2, dplyr, textshape, numform, tidyr, lsa, viridis)
## get the geoms
geoms <- gsub('^geom_', '', grep('^geom_', p_funs(ggplot2), value = TRUE))
## function to grab the aesthetics
get_aesthetics <- function(name, type){
obj <- switch(type, geom = ggplot2:::find_subclass("Geom", name, globalenv()),
stat = ggplot2:::find_subclass("Stat", name, globalenv()))
aes <- ggplot2:::rd_aesthetics_item(obj)
req <- obj$required_aes
data_frame(
aesthetic = union(req, sort(obj$aesthetics())),
required = aesthetic %in% req,
)
}
## loop through and grab the aesthetics per geom
aesthetics_list <- lapply(geoms, function(name){
type <- 'geom'
name <- switch(name,
jitter = 'point',
freqpoly = 'line',
histogram = 'bar',
name
)
out <- try(get_aesthetics(name, 'geom'), silent = TRUE)
if (inherits(out, 'try-error')) out %
setNames(geoms)
## convert the list of data.frames to one tidy data.frame
aesthetics %
tidy_list('geom') %>%
tbl_df()
aesthetics
geom aesthetic required
1 abline slope TRUE
2 abline intercept TRUE
3 abline alpha FALSE
4 abline colour FALSE
5 abline group FALSE
6 abline linetype FALSE
7 abline size FALSE
8 area x TRUE
9 area y TRUE
10 area alpha FALSE
# ... with 341 more rows
Geoms: Getting Acquainted
First I wanted to get to know geoms again. There are currently 44 of them.
length(unique(aesthetics$geom)) ## 44
This bar plot details the geoms and how many aesthetics are optional/required.
geom_ord %
count(geom) %>%
arrange(n) %>%
pull(geom)
aesthetics %>%
mutate(geom = factor(geom, levels = geom_ord)) %>%
ggplot(aes(geom, fill = required)) +
geom_bar() +
coord_flip() +
scale_y_continuous(expand = c(0, 0), limits = c(0, 15)) +
scale_fill_manual(name = 'Required', values = c('gray88', '#1C86EE'), labels = f_response) +
theme_minimal() +
theme(
panel.grid.major.y = element_blank(),
axis.text = element_text(color = 'grey55', size = 10),
legend.key.size = unit(.35, 'cm'),
legend.title = element_text(color = 'grey30', size = 10),
legend.position = c(.76, .1),
axis.title = element_text(color = 'gray55')
) +
labs(
x = 'Geom',
y = 'Count',
title = 'Count of Geom Aesthetics',
subtitle = 'Distribution of geom aesthetic count filled by requirement flag.'
)
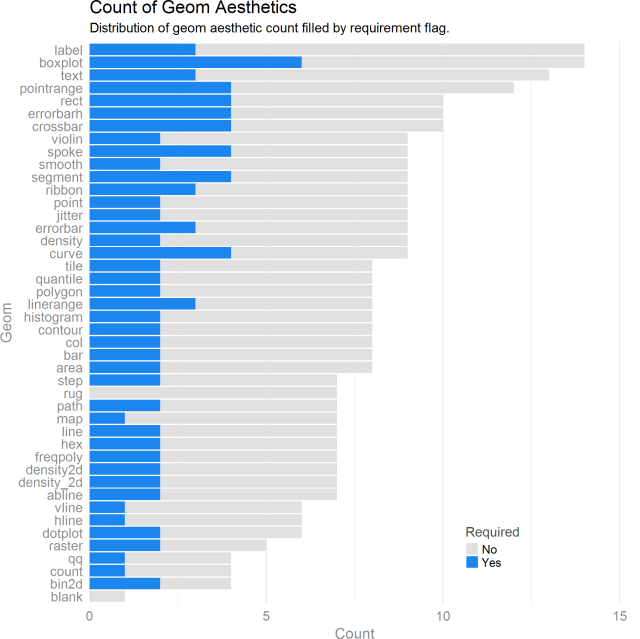
Some interesting things come out. Most geoms have 2ish required aesthetics. The boxplot geom has the most required and unrequired aesthetics. Sensibly, a blank geom requires no aesthetics. I wanted to see what all of these aesthetics were for the boxplot. Some quick dplyr-ing has us there in no time.
aesthetics %>%
filter(geom == 'boxplot')
geom aesthetic required
1 boxplot x TRUE
2 boxplot lower TRUE
3 boxplot upper TRUE
4 boxplot middle TRUE
5 boxplot ymin TRUE
6 boxplot ymax TRUE
7 boxplot alpha FALSE
8 boxplot colour FALSE
9 boxplot fill FALSE
10 boxplot group FALSE
11 boxplot linetype FALSE
12 boxplot shape FALSE
13 boxplot size FALSE
14 boxplot weight FALSE
Seems that x is the only aesthetic that is truly required. The other “required” ones are computed if you just supply x.
Aesthetics: May I join You?
Now time to get to know aesthetics. Are there others like stroke that I’ve overlooked? There are 36 aesthetics currently. I see right away that there is a weight aesthetic I’ve never seen before.
length(unique(aesthetics$aesthetic)) ## 36
aes_ord %
count(aesthetic) %>%
arrange(n) %>%
pull(aesthetic)
aesthetics %>%
mutate(aesthetic = factor(aesthetic, levels = aes_ord)) %>%
ggplot(aes(aesthetic, fill = required)) +
geom_bar() +
scale_y_continuous(expand = c(0, 0), limits = c(0, 45), breaks = seq(0, 45, by = 5)) +
scale_fill_manual(name = 'Required', values = c('gray88', '#1C86EE'), labels = f_response) +
theme_minimal() +
theme(
panel.grid.major.y = element_blank(),
panel.grid.minor = element_blank(),
axis.text = element_text(color = 'grey55', size = 10),
legend.key.size = unit(.35, 'cm'),
legend.title = element_text(color = 'grey30', size = 10),
legend.position = c(.83, .1),
axis.title = element_text(color = 'gray55')
) +
labs(
x = 'Aesthetic',
y = 'Count',
title = 'Count of Aesthetics',
subtitle = 'Distribution of aesthetics filled by requirement flag.'
) +
coord_flip()
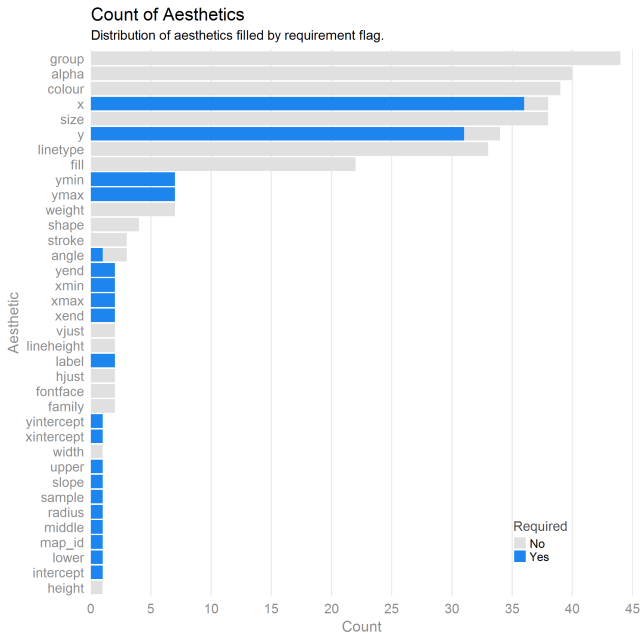
It seems from this plot that almost every geom requires an x/y position aesthetic. That makes sense. What doesn’t make sense are cases besides geom_blank that don’t require x/y.
aesthetics %>%
filter(aesthetic %in% c('x', 'y') & !required)
geom aesthetic required
1 count y FALSE
2 qq x FALSE
3 qq y FALSE
4 rug x FALSE
5 rug y FALSE
This dplyr summarize/filter shows where an x/y are possible aesthetics but not required. Sensible. But are there some geoms that x/y aren’t even in their mapping?
aesthetics %>%
group_by(geom) %>%
summarize(
has_x = 'x' %in% aesthetic,
has_y = 'y' %in% aesthetic
) %>%
filter(!has_x | ! has_y) %>%
arrange(has_x)
geom has_x has_y
1 abline FALSE FALSE
2 blank FALSE FALSE
3 hline FALSE FALSE
4 map FALSE FALSE
5 rect FALSE FALSE
6 vline FALSE FALSE
7 boxplot TRUE FALSE
8 errorbar TRUE FALSE
9 linerange TRUE FALSE
10 ribbon TRUE FALSE
Dig into the documentation if you want to make sense of why these geoms don’t require x/y positions.
Geoms & Aesthetics Intersect
OK so we’ve explored variability in geoms and aesthetics a bit…let’s see how they covary. The heatmap below provides an understanding of what geoms utilize what aesthetics and if they are required. NA means that the aesthetic is not a part of the geom’s mapping.
boolean %
count(geom, aesthetic) %>%
spread(aesthetic, n, fill = 0)
boolean %>%
gather(aesthetic, value, -geom) %>%
left_join(aesthetics, by = c('geom', 'aesthetic')) %>%
mutate(
aesthetic = factor(aesthetic, levels = rev(aes_ord)),
geom = factor(geom, levels = rev(geom_ord))
) %>%
ggplot(aes(y = aesthetic, x = geom, fill = required)) +
geom_tile(color = 'grey95') +
scale_fill_manual(name = 'Required', values = c('gray88', '#1C86EE'), labels = f_response) +
theme_minimal() +
theme(
axis.ticks = element_blank(),
axis.text.y = element_text(size = 8, margin = margin(r = -3)),
axis.text.x = element_text(size = 8, hjust = 1, vjust = 1,
angle = 45, margin = margin(t = -3)),
legend.key.size = unit(.35, 'cm'),
legend.title = element_text(color = 'grey30', size = 10),
panel.grid = element_blank(),
axis.title = element_text(color = 'gray55')
) +
labs(
title = 'Geom & Aesthetic Co-occurrence',
subtitle = NULL,
y = 'Aesthetic',
x = 'Geom'
)
Here we can see that weight aesthetic again. Hmmm, boxplot has it and other, mostly univariate functions as well. The documentation is a bit sparse on it. Hadley says: https://github.com/tidyverse/ggplot2/issues/1893
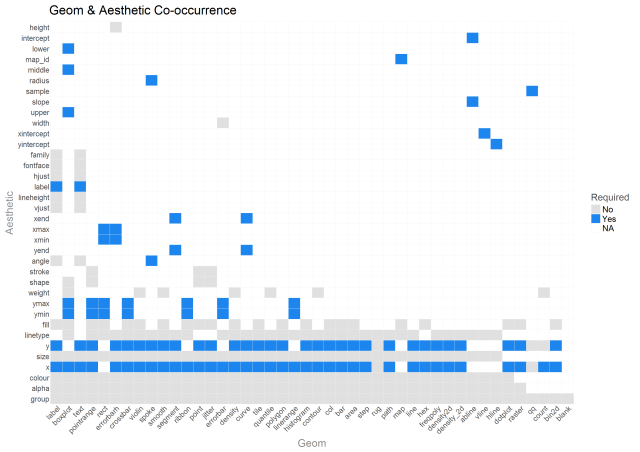
Also there are two geoms that are just now catching my eye…geom_count & geom_spoke. I’ll come back to them later.
Also, there are a few aesthetics like height and intercept that are one time use only aesthetics. Also, the bottom 10 aesthetics are used, by far, the most frequently. For beginners, these are the ones that will really pay to learn quickly.
Geom Similarity
I thought it my be fun to use the geoms aesthetics to see if we could cluster aesthetically similar geoms closer together. The heatmap below uses cosine similarity and heirarchical clustering to reorder the matrix that will allow for like geoms to be found closer to one another (note that today I learned from “R for Data Science” about the seriation package [https://cran.r-project.org/web/packages/seriation/index.html] that may make this matrix reordering task much easier).
boolean %>%
column_to_rownames() %>%
as.matrix() %>%
t() %>%
cosine() %>%
cluster_matrix() %>%
tidy_matrix('geom', 'geom2') %>%
mutate(
geom = factor(geom, levels = unique(geom)),
geom2 = factor(geom2, levels = unique(geom2))
) %>%
group_by(geom) %>%
ggplot(aes(geom, geom2, fill = value)) +
geom_tile() +
scale_fill_viridis(name = bquote(cos(theta))) +
theme(
axis.text.y = element_text(size = 8) ,
axis.text.x = element_text(size = 8, hjust = 1, vjust = 1, angle = 45),
legend.position = 'bottom',
legend.key.height = grid::unit(.2, 'cm'),
legend.key.width = grid::unit(.7, 'cm'),
axis.title = element_text(color = 'gray55')
) +
labs(
title = "ggplot2 Geom Cosine Similarity",
subtitle = 'Geoms with similar aesthetics are clustered together along the diagonal.',
x = 'Geom',
y = 'Geom'
)
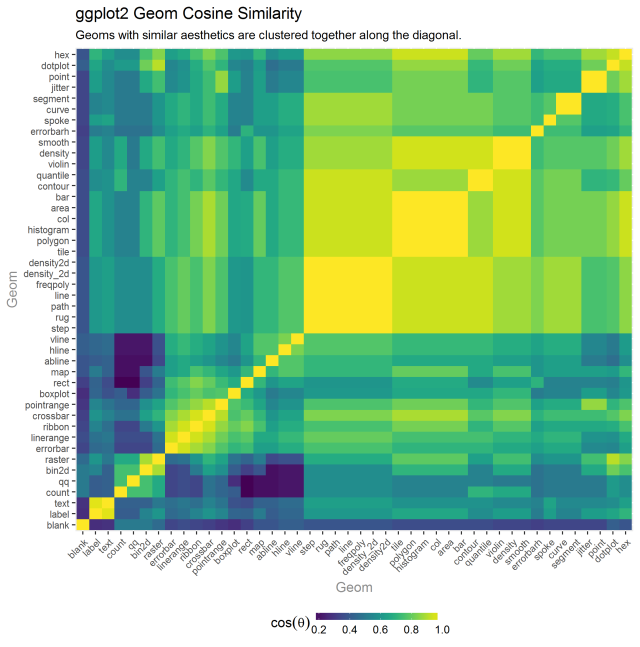
Looking at the bright square clusters along the diagonal is a good starting place for understanding which geoms tend to aesthetically cluster together. Generally, this ordering seems pretty reasonable.
Aesthetic Similarity
I performed the same analysis of aesthetics, asking which ones tended to be used within the same geoms.
boolean %>%
column_to_rownames() %>%
as.matrix() %>%
cosine() %>%
{x <- .; diag(x) %
cluster_matrix() %>%
tidy_matrix('aesthetic', 'aesthetic2') %>%
mutate(
aesthetic = factor(aesthetic, levels = unique(aesthetic)),
aesthetic2 = factor(aesthetic2, levels = unique(aesthetic2))
) %>%
group_by(aesthetic) %>%
ggplot(aes(aesthetic, aesthetic2, fill = value)) +
geom_tile() +
scale_fill_viridis(name = bquote(cos(theta))) +
theme(
axis.text.y = element_text(size = 8) ,
axis.text.x = element_text(size = 8, hjust = 1, vjust = 1, angle = 45),
legend.position = 'bottom',
legend.key.height = grid::unit(.2, 'cm'),
legend.key.width = grid::unit(.7, 'cm'),
axis.title = element_text(color = 'gray55')
) +
labs(
title = "ggplot2 Aesthetics Cosine Similarity",
subtitle = f_wrap(c(
'Aesthetics that tend to be used together within the same geoms are clustered together along the diagonal.'
), width = 95, collapse = TRUE
),
x = 'Aesthetic',
y = 'Aesthetic'
)
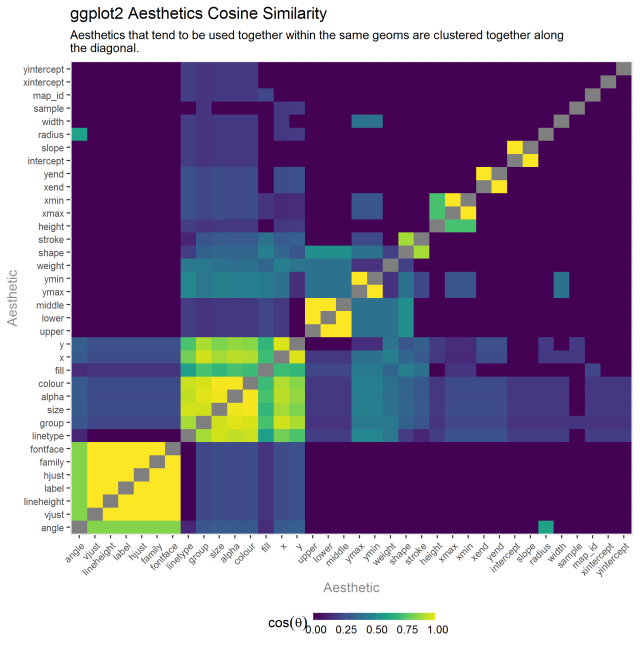
The result is pretty sensible. The lower left corner has the largest cluster which seems to be related to text based geoms. The next cluster up and to the right one, has group, size, x, y, etc. This seems to be the most common set of typically geometric aesthetics. The upper, lower, middle cluster is specific to the boxplot summary stat. Stroke and shape as a cluster are related to geoms that are point based.
Geom Similarity: Required Aesthetics
The last clustering activity I wanted was to reduce the seahetics to jsut required (as we might assume these are the truest attributes of a geom) and see which geoms cluster from that analysis.
boolean2 %
filter(required) %>%
count(geom, aesthetic) %>%
spread(aesthetic, n, fill = 0)
boolean2 %>%
column_to_rownames() %>%
as.matrix() %>%
t() %>%
cosine() %>%
cluster_matrix() %>%
tidy_matrix('geom', 'geom2') %>%
mutate(
geom = factor(geom, levels = unique(geom)),
geom2 = factor(geom2, levels = unique(geom2))
) %>%
group_by(geom) %>%
ggplot(aes(geom, geom2, fill = value)) +
geom_tile() +
scale_fill_viridis(name = bquote(cos(theta))) +
theme(
axis.text.y = element_text(size = 8) ,
axis.text.x = element_text(size = 8, hjust = 1, vjust = 1, angle = 45),
legend.position = 'bottom',
legend.key.height = grid::unit(.2, 'cm'),
legend.key.width = grid::unit(.7, 'cm'),
axis.title = element_text(color = 'gray55')
) +
labs(
title = "ggplot2 Geom Cosine Similarity",
subtitle = 'Geoms with similar aesthetics are clustered together along the diagonal.',
x = 'Geom',
y = 'Geom'
)
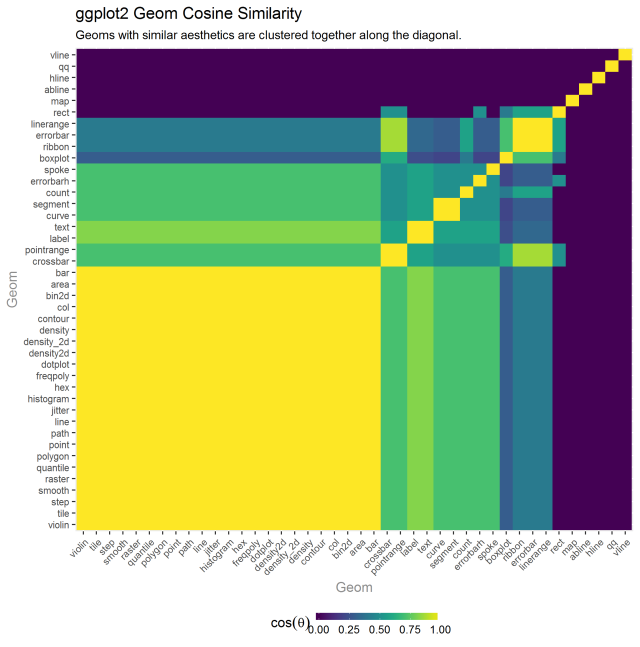
This seemed less interesting. I didn’t really have a plausible explanation for what patterns did show up and for the most part, clusters became really large or really small. I’m open to others’ interpretations.
A Few Un-/Re-discovered ggplot2 Features
I did want to learn more about geom_count & geom_spoke in the remaining exploration.
?geom_count
ggplot(diamonds, aes(x = cut, y = clarity)) +
geom_count(aes(size = ..prop.., group = 1)) +
scale_size_area(max_size = 10)
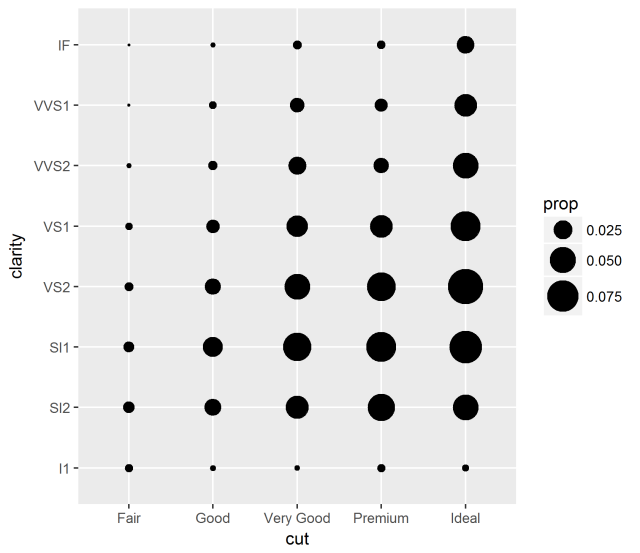
Oh yeah! Now I remember. A shortcut to make the geom_point bubble plot for investigating categorical variable covariance as an alternative to the heatmap.
?geom_spoke
df <- expand.grid(x = 1:10, y=1:10)
df$angle <- runif(100, 0, 2*pi)
df$speed <- runif(100, 0, sqrt(0.1 * df$x))
ggplot(df, aes(x, y)) +
geom_point() +
geom_spoke(aes(angle = angle), radius = 0.5)
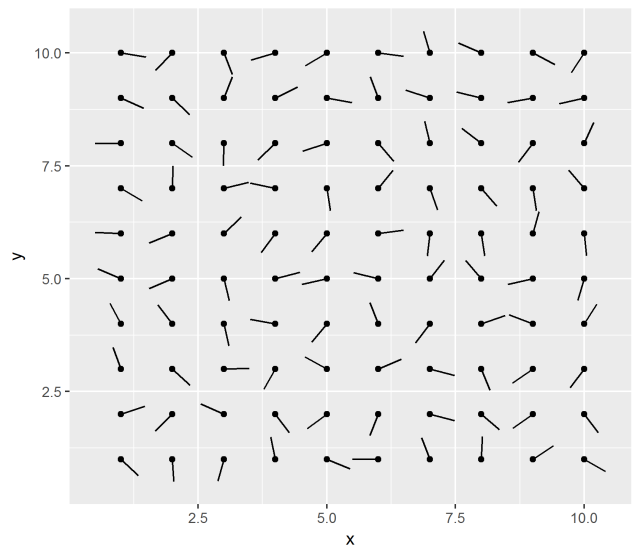
Interesting. The documentation says:
…useful when you have variables that describe direction and distance.
Not sure if I have a use case for my own work. But I’ll store it in the vault (but better than I remembered geom_count).
Learning More About Geoms & Aesthetics
I wanted to leave people with a quick reference guide that RStudio has kindly provided to help give quick reference to geoms and aesthetics and whe to use them.